My research is about creating useful methods for biological applications and finding hidden relationships in biological data using statistical methods and deep learning.
Bryant P., Noé F. Structure prediction of alternative protein conformations. Preprint (2023)
Protein complex prediction
Bryant P., Noé F. Improved protein complex prediction with AlphaFold-multimer by denoising the MSA profile. Preprint (2023)
Bryant, P., Pozzati, G., Zhu, W. et al. Predicting the structure of large protein complexes using AlphaFold and Monte Carlo tree search. Nat Commun 13, 6028 (2022).
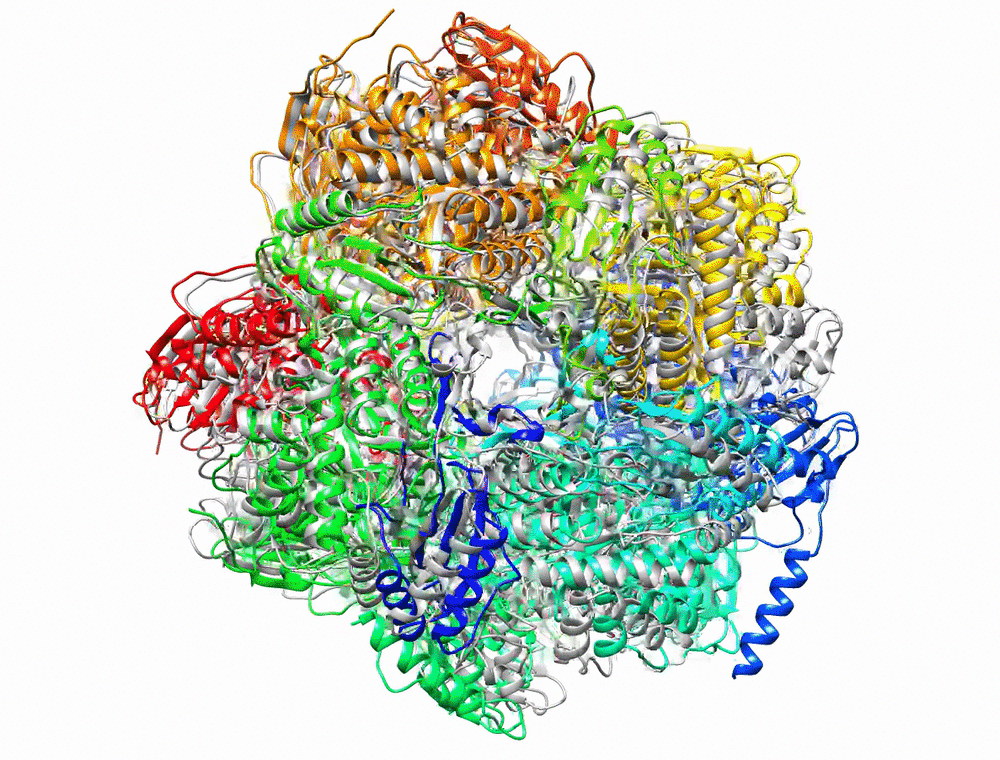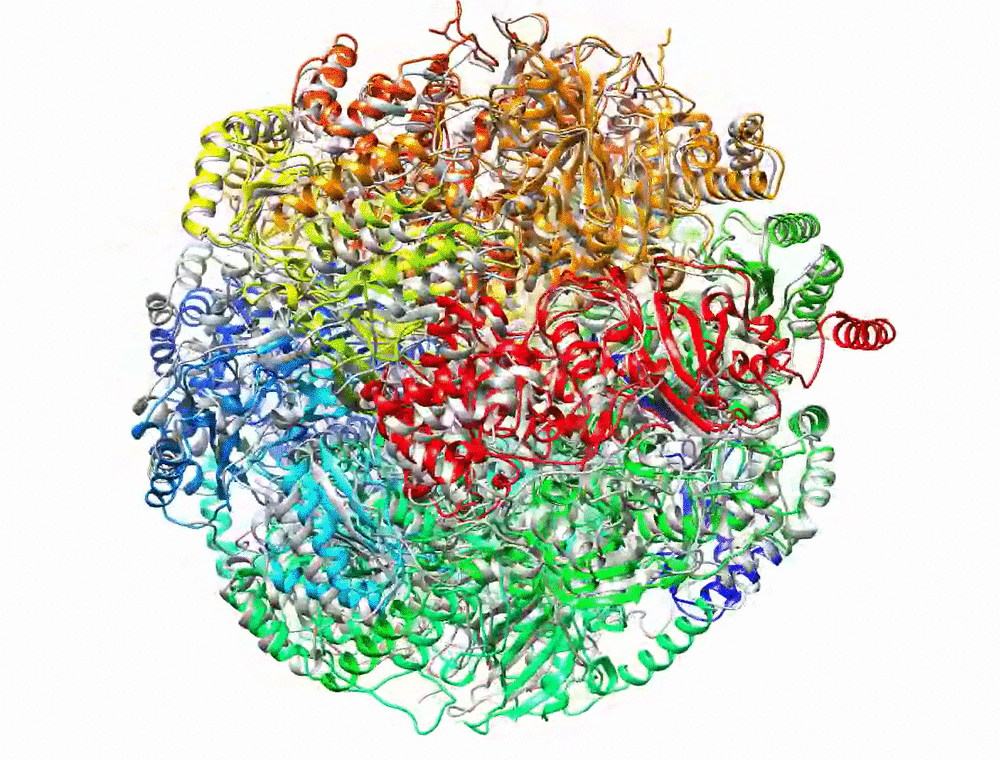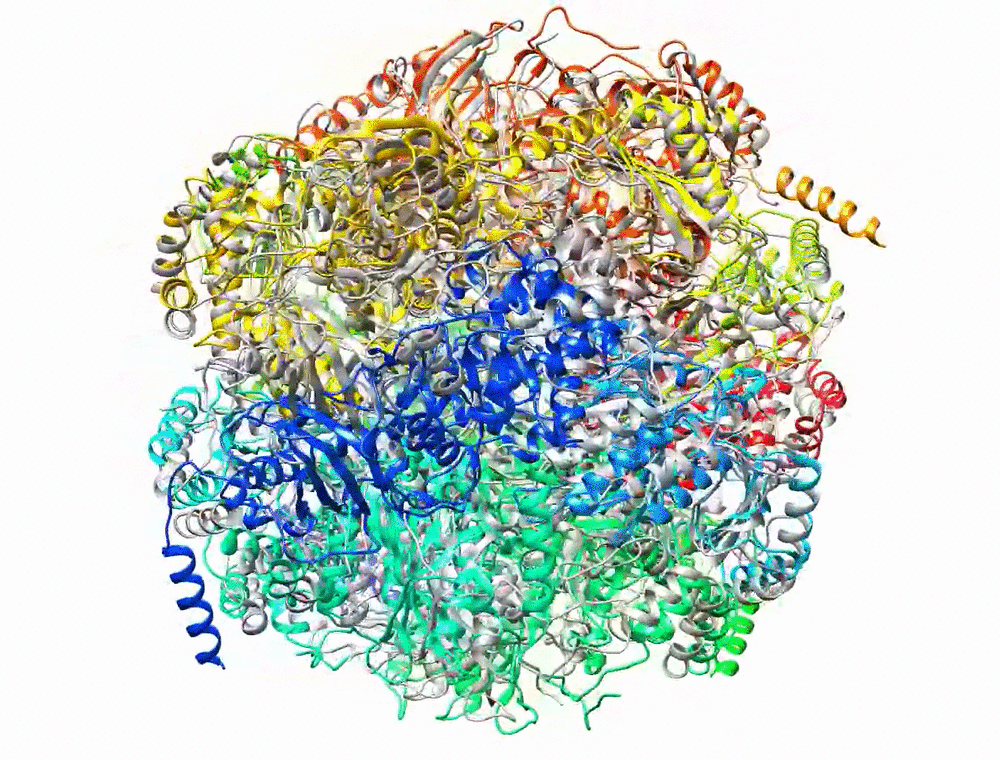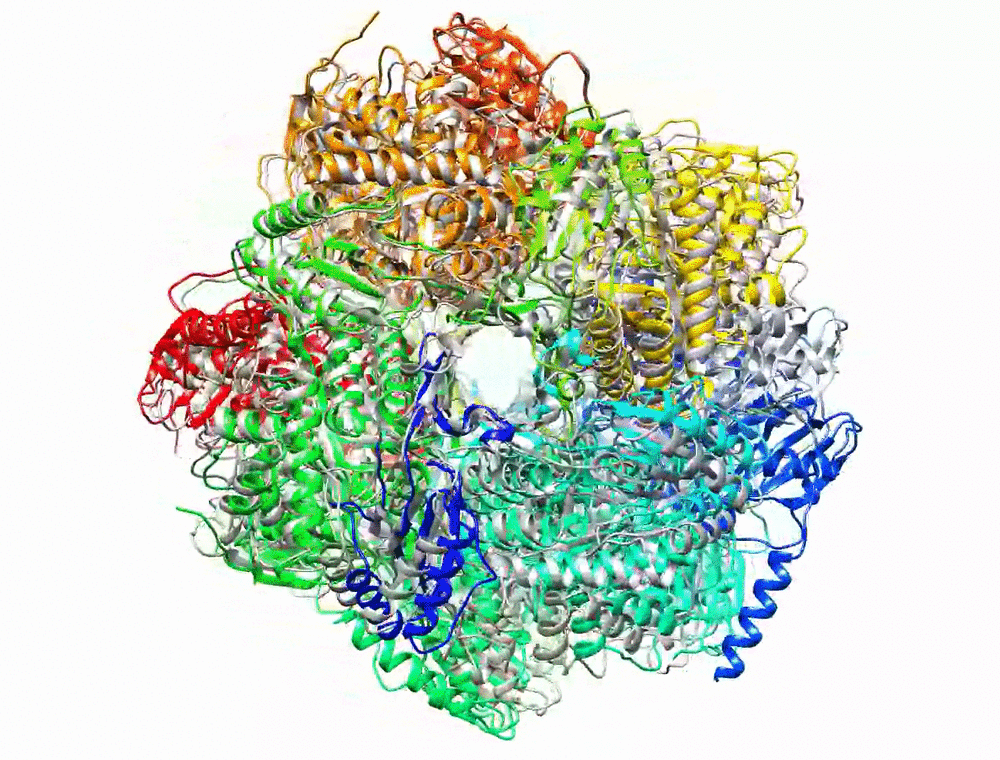
Bryant P, Pozzati G, Elofsson A. Improved prediction of protein-protein interactions using AlphaFold2. Nat Commun. 2022;13: 1–11.
Burke, D.F., Bryant, P., Barrio-Hernandez, I. et al. Towards a structurally resolved human protein interaction network. Nat Struct Mol Biol (2023). https://doi.org/10.1038/s41594-022-00910-8
Akdel M, Pires DEV, Pardo EP, Jänes J, Zalevsky AO, Mészáros B, Bryant P et al. A structural biology community assessment of AlphaFold2 applications. Nat Struct Mol Biol 29, 1056–1067 (2022). https://doi.org/10.1038/s41594-022-00849-w
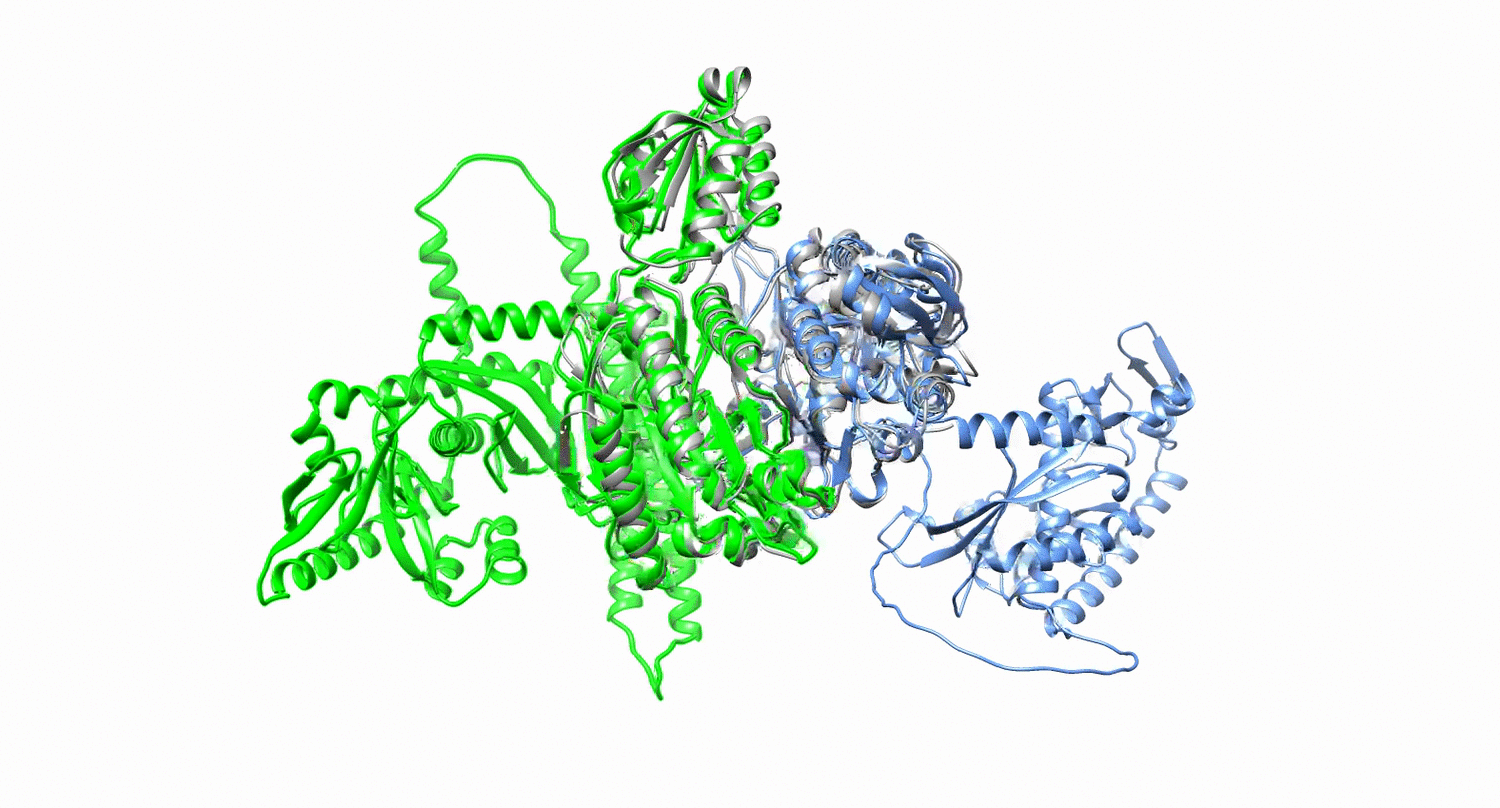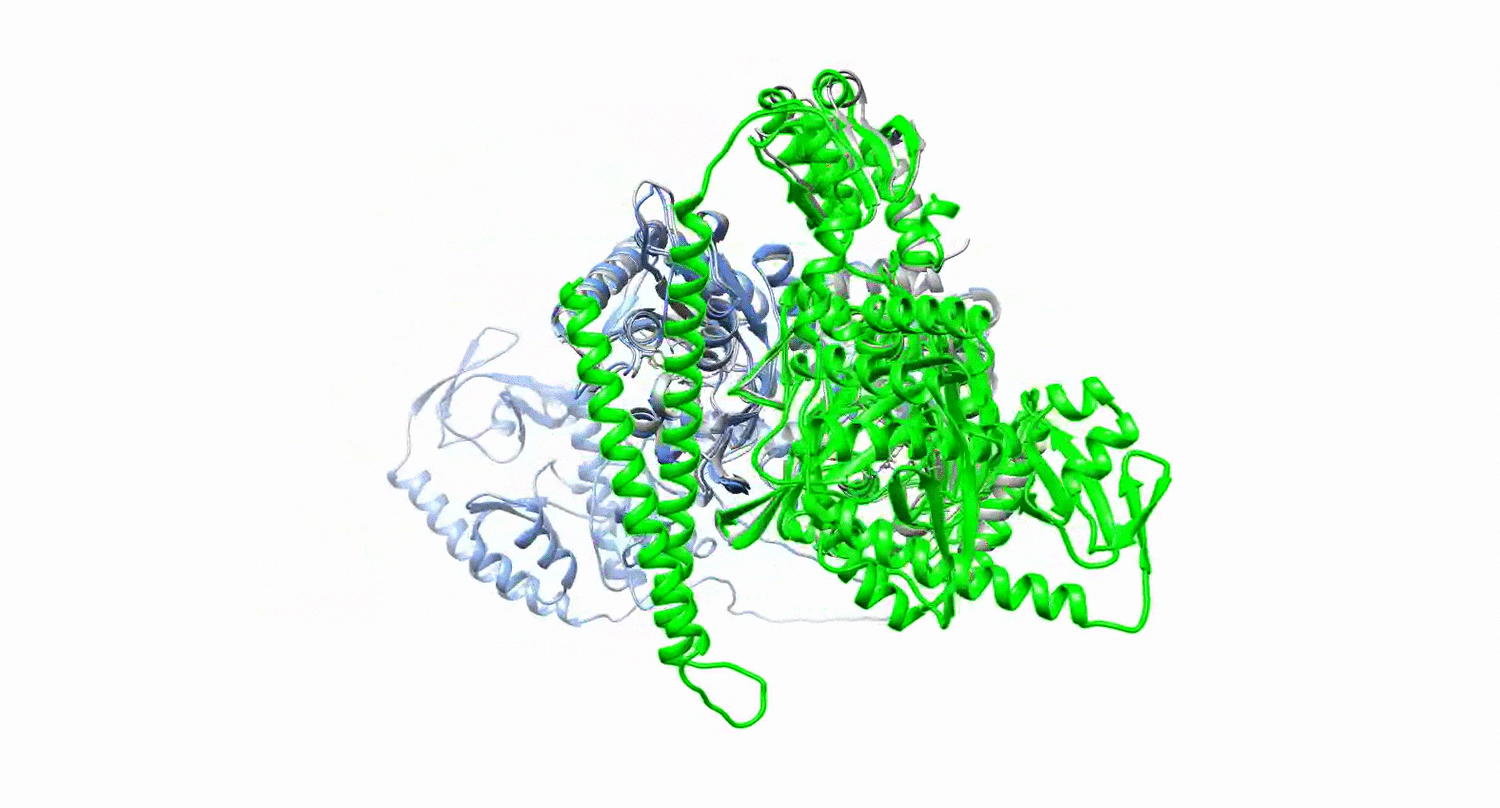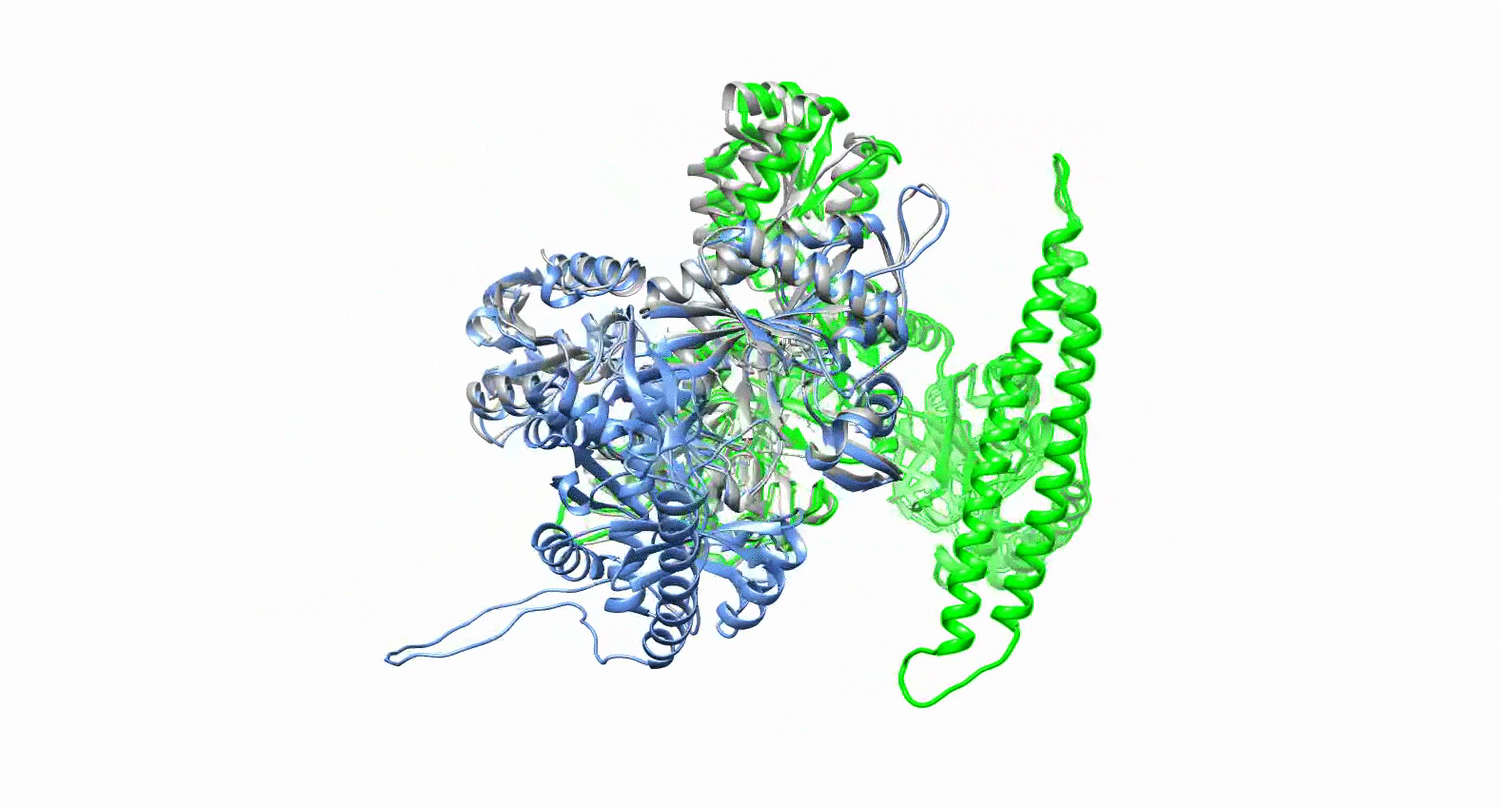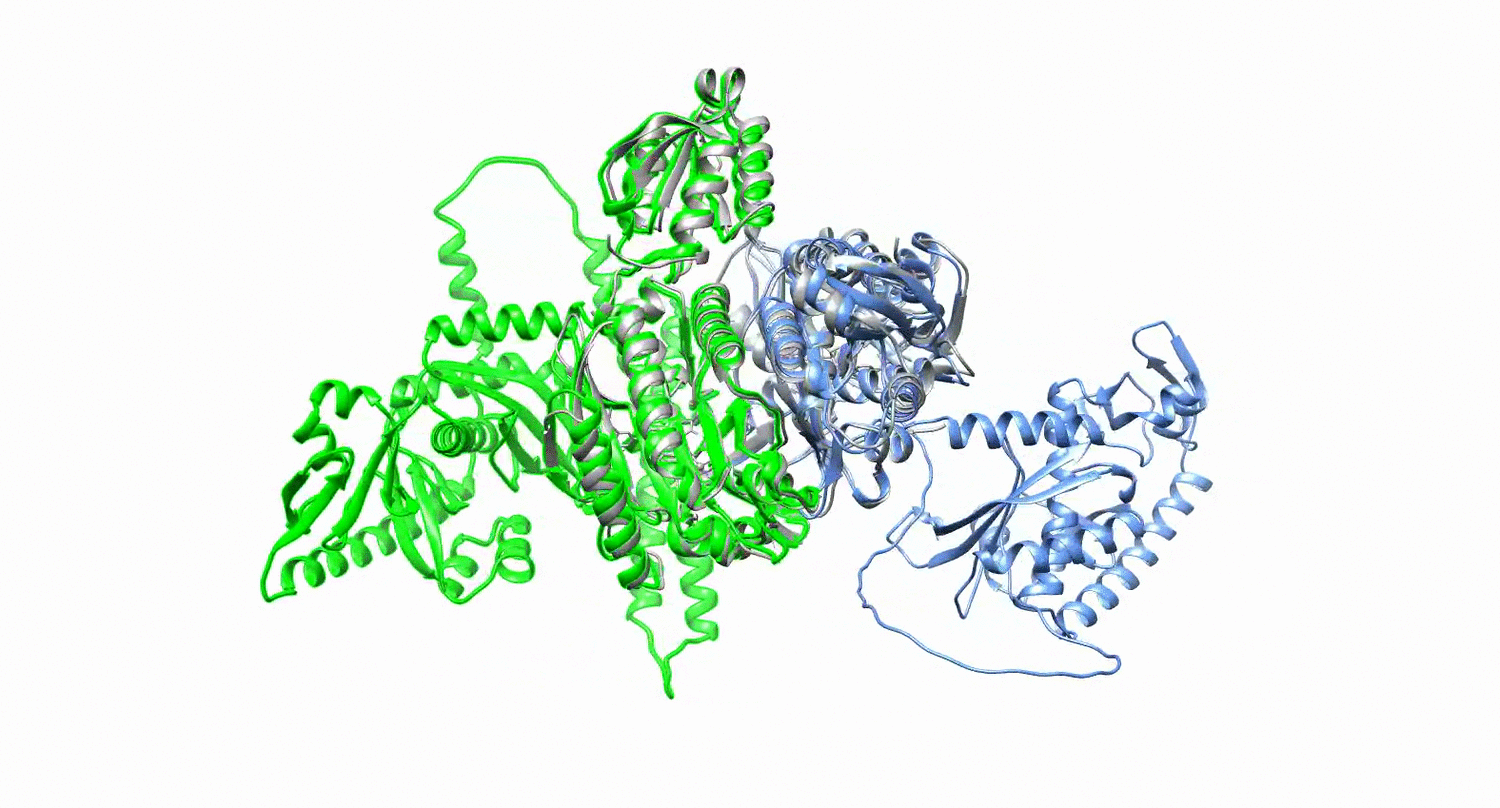
Protein design
Bryant P, Elofsson A. EvoBind: in silico directed evolution of peptide binders with AlphaFold. bioRxiv. 2022. p. 2022.07.23.501214. doi:10.1101/2022.07.23.501214
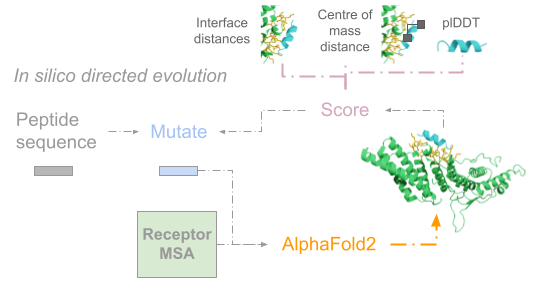
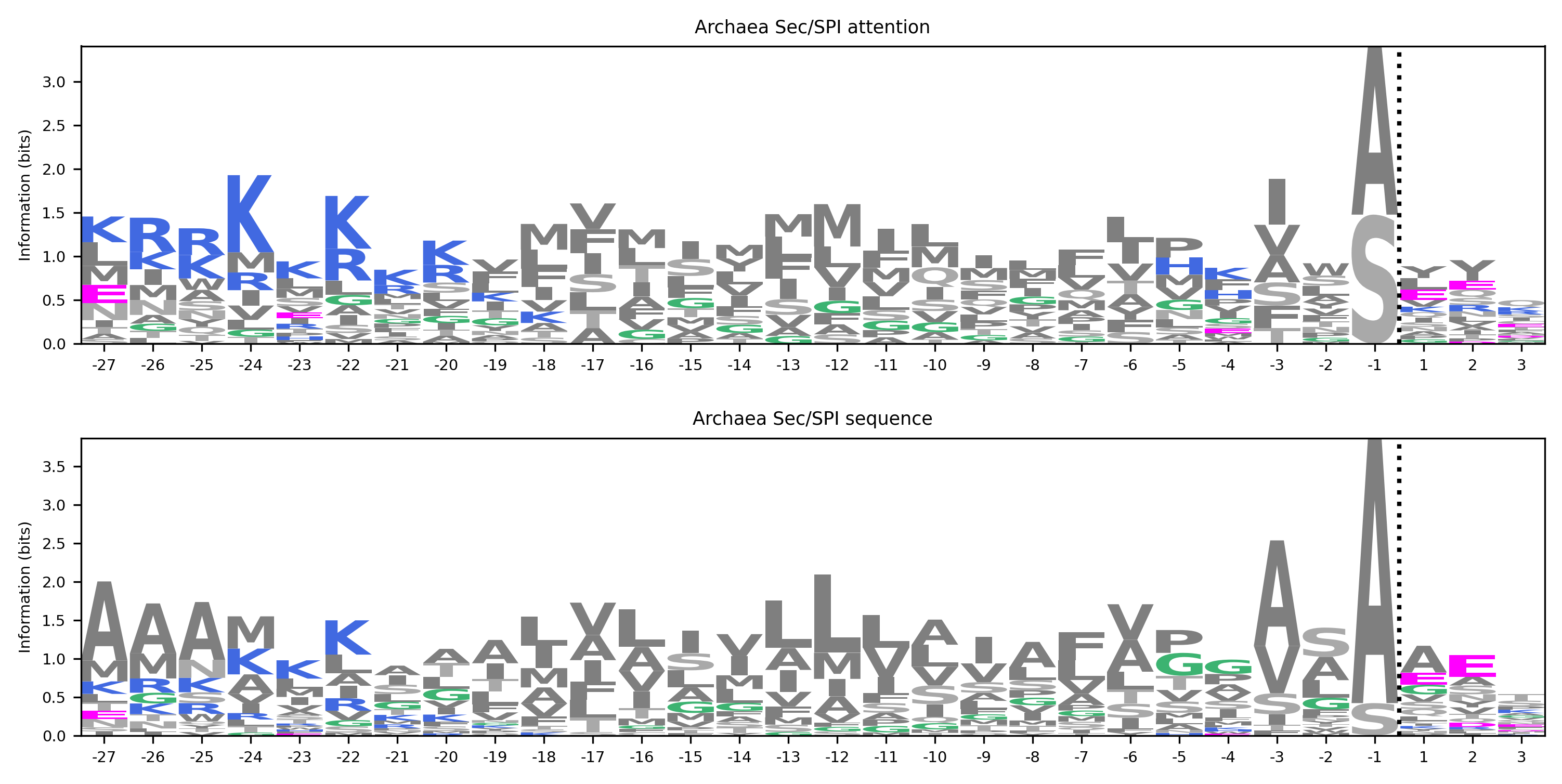
Signal peptides
Bryant P, Elofsson A. Studying signal peptides with attention neural networks informs cleavage site predictions. MLSB Workshop at NeurIPS 2021
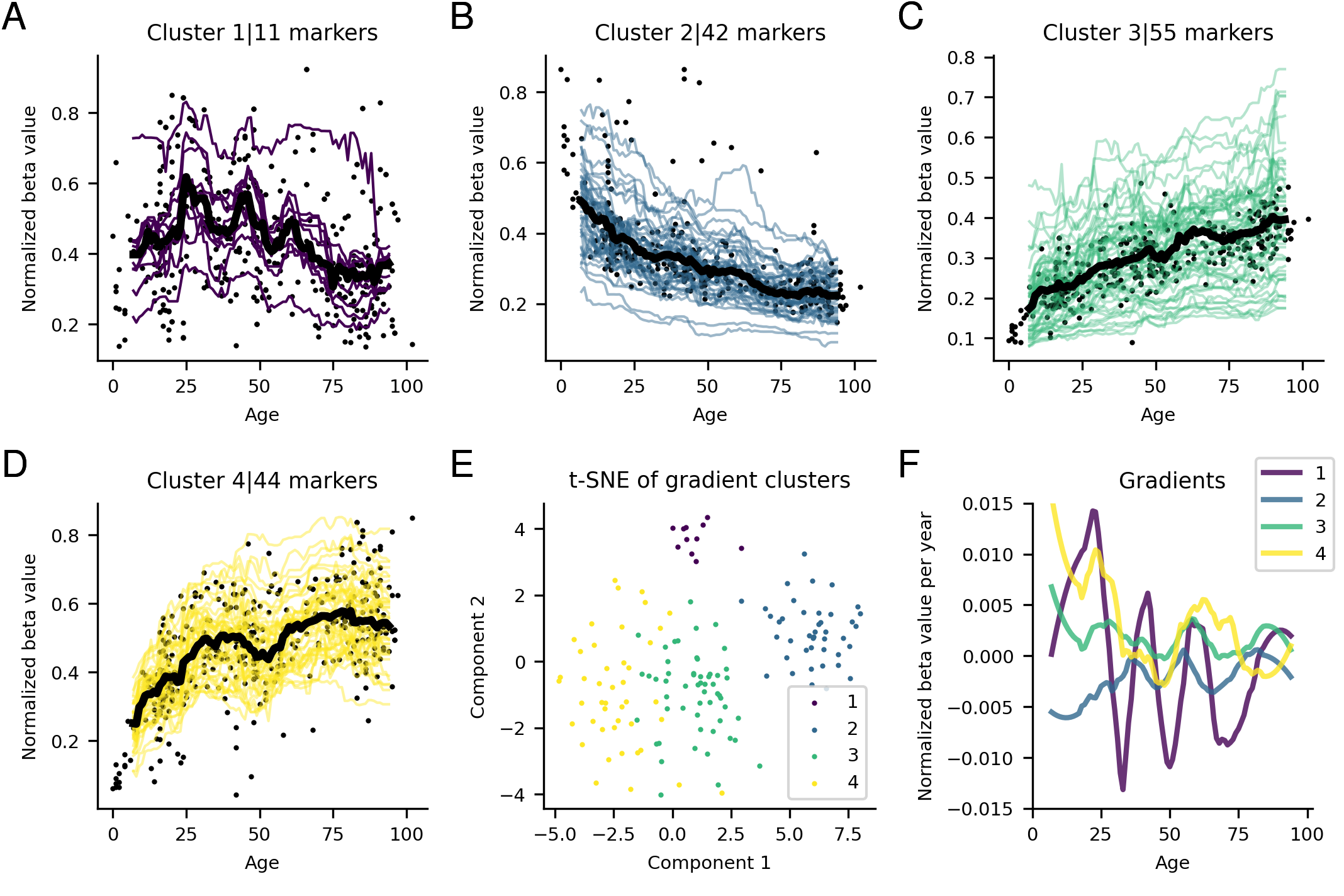
Epigenetics and ageing
Bryant P, Elofsson A. 2021. The relationship between ageing and changes in the human blood and brain methylomes. NAR Genomics and Bioinformatics, Volume 4, Issue 1, March 2022
COVID-19 (SARS-CoV-2)
Bryant P, Elofsson A. 2020. Estimating the impact of mobility patterns on COVID-19 infection rates in 11 European countries. PeerJ 8:e9879 https://doi.org/10.7717/peerj.9879
Bryant P, Elofsson A. 2020. The limits of estimating COVID-19 intervention effects using Bayesian models (preprint). medRxiv
Protein Evolution
Bryant P, Elofsson A. 2020. Decomposing Structural Response Due to Sequence Changes in Protein Domains with Machine Learning. Journal of Molecular Biology, Volume 432, Issue 16 https://doi.org/10.1016/j.jmb.2020.05.021

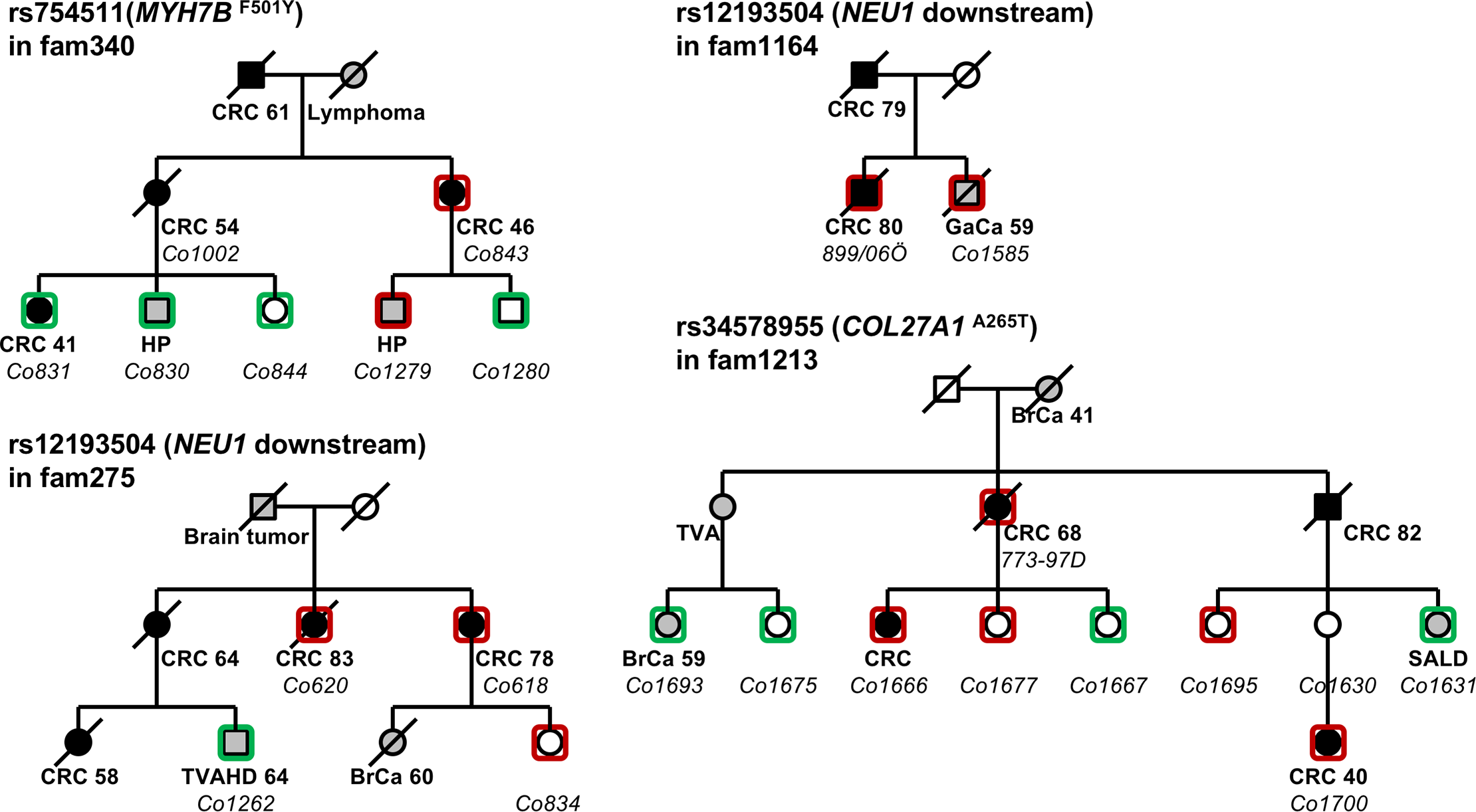
Familial Cancer
A Swedish Genome-Wide Haplotype Association Analysis Identifies a Novel Breast Cancer Susceptibility Locus in 8p21.2 and Characterizes Three Loci on Chromosomes 10, 11 and 16
Linkage Analysis Revealed Risk Loci on 6p21 and 18p11.2-q11.2 in Familial Colon and Rectal Cancer, Respectively.” European Journal of Human Genetics: EJHG 27 (8): 1286–95.
Recurrent, Low-Frequency Coding Variants Contributing to Colorectal Cancer in the Swedish Population. PloS One 13 (3): e0193547.
Code for analysis of Next Generation Sequencing data.






